my_loocv <- function(fit){
dat <- fit$model
y <- all.vars(fit$call)[1]
errors <- vector(mode = "numeric", length = nrow(dat))
for(i in 1:nrow(dat)){
to_pred_i <- dat[i, ]
fit_no_i <- update(fit, data = dat[-i, ])
pred_i <- predict(fit_no_i, to_pred_i)
errors[i] <- pred_i - to_pred_i[1, y]
}
return(errors)
}
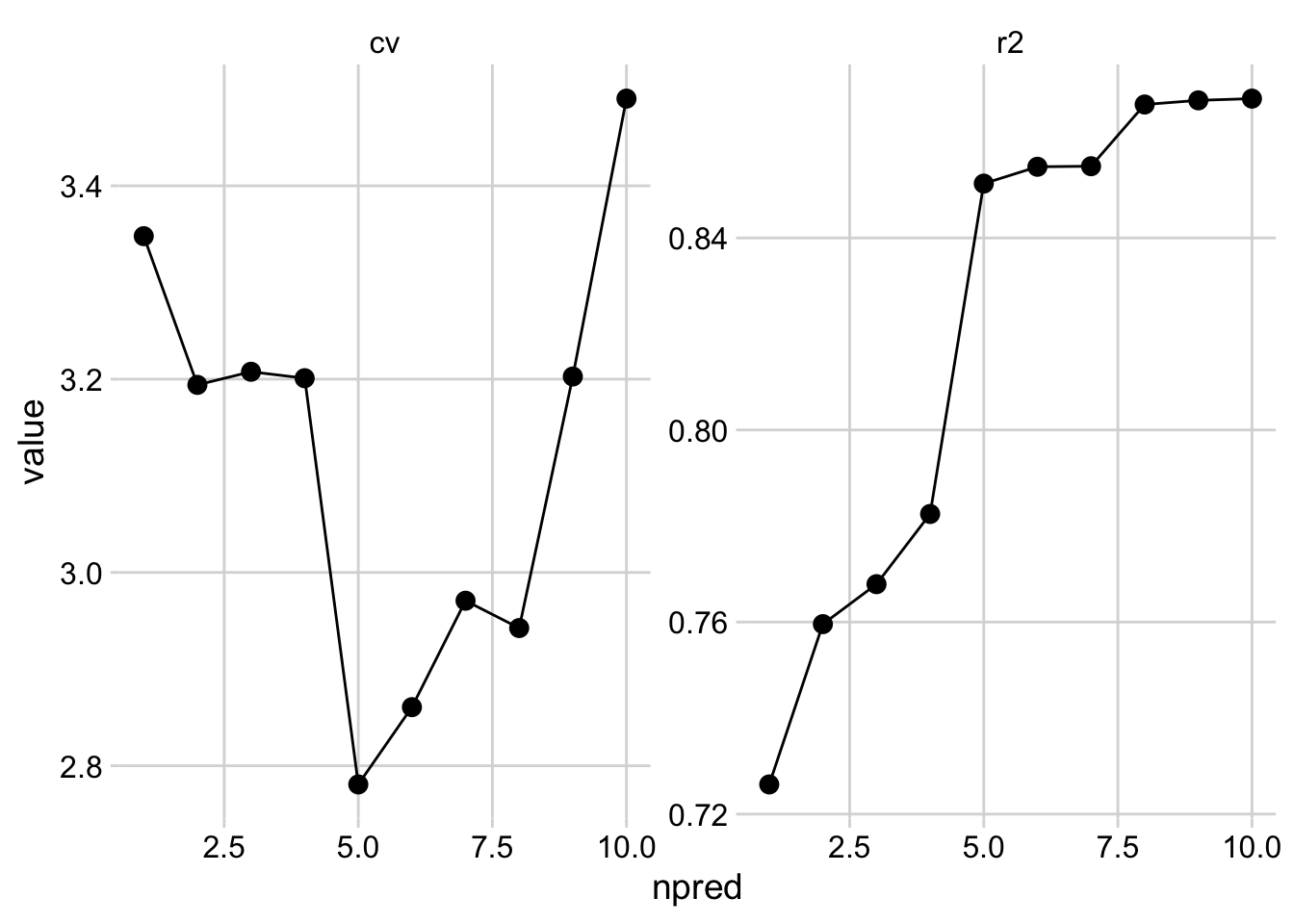
cv <- map_dbl(fit_list, function(x) get_rmse(my_loocv(x))) # get loo-cv mean error
npred <- map_dbl(fit_list, function(i) length(all.vars(i$call))-2) # get number of predictors
r2 <- map_dbl(fit_list, function(mod) summary(mod)$r.squared) # get rsquared from fitted models
loo_cv <- data.frame(
cv, r2, npred
)
# Plotting
loo_cv %>%
tidyr::pivot_longer(c(1,2), names_to = "measure", values_to = "value") %>%
ggplot(aes(x = npred, y = value)) +
geom_line() +
geom_point(size = 3) +
facet_wrap(~measure, scales = "free") +
cowplot::theme_minimal_grid()